Role of PA5003 in Colistin-induced Over-expression of Pseudomonas Quinolone Signal in P. aeruginosa
Shuya Zhai
James Schafhauser
Geoffrey McKay
Dao Nguyen*
McGill University
Abstract
Cystic fibrosis (CF) is an autosomal recessive disorder caused by mutations in the cystic fibrosis transmembrane regulator (CFTR) gene. CF patients are more susceptible to opportunistic infections by different bacteria, and most importantly P. aeruginosa,which causes both acute and chronic infections because of its ability to secrete numerous virulent compounds and degradative enzymes. Expression of the virulent compounds is controlled by quorum sensing. The Pseudomonas Quinolone Signal system (PQS) is one of three quorum-sensing networks in P. aeruginosa. A study found that sub-inhibitory concentration of the cationic antimicrobial peptide (CAMP) colistin induced PQS overproduction in P. aeruginosa, but the exact mechanism remains elusive. It was also previously demonstrated that the gene PA5003 is required for P. aeruginosa to recognize and respond to the presence of the CAMPs (colistin). From a random mutagenesis screen conducted, our lab also demonstrated that loss of PA5003 led to reduced PQS expression. These observations led us to hypothesize that PA5003 is required for colistin-induced PQS over-production. In this study, we confirmed that colistin induced over-expression of pqsA, one of the PQSbiosynthesis genes, and demonstrated that PA5003 plays a role in the overexpression of pqsA induced by sub-inhibitory concentration of colistin.
Introduction
Cystic fibrosis (CF) is an autosomal recessive genetic disorder caused by mutations in the cystic transmembrane regulator (CFTR) gene. It mostly affects the lungs, but also other organs including pancreas, liver, and intestine, and is characterized by abnormal transport of chloride and sodium ions across the epithelium, leading to thick, viscous secretions.1 CF is the most common fatal genetic disease affecting Canadian children and young adults. It is estimated that one in every 3,600 children born in Canada are affected by CF.
CF patients are more susceptible to opportunistic infections by different bacteria. Among them, P. aeruginosa is the most dominant pathogen.2 P. aeruginosa causes both acute and chronic infections with its ability to secrete numerous virulent compounds and degradative enzymes.2 In P. aeruginosa, the expression and secretion of numerous virulence factors are regulated by a hierarchical quorum sensing system that is a cell-to-cell communication system and mediated by the two chemically distinct classes of signal molecules, the N-acylhomoserine lactones and the 4-hydroxy-2-akylquinolone (HAQ).3 The latter group consists of more than 50 compounds and includes the primary signal molecule 2-heptyl-3-hydroxy-4-quinolone that is commonly referred to as the pseudomonas quinolone signal (PQS).4 PQS belongs to a group of structurally related compounds called 4-hydroxy-2-akylquinolone (HAQ).5 HAQs are synthesized by the pqsABCDE operon as well as pqsH and pqsL. In addition, pqsR is a transcriptional regulator that binds with PQS and up-regulates the expression of pqsA. pqsH is responsible for the conversion of HHQ (2-heptyl-4-quinolone) into PQS. No HAQ molecules are produced if any of pqsA, pqsAB, pqsC or pqsD genes are deleted.
The stringent response is a bacterial stress response, which responds to amino-acid starvation, fatty acid limitation, iron limitation, heat shock, and several other environmental stresses.6 The stringent response is mediated by (p)ppGpp (penta and tetra-phosphorylated guanosine). In Gram-negative bacteria, (p)ppGpp is synthesized by RelA (a ribosome-dependent synthetase) and SpoT (a bifunctional synthetase and hydrolase), and it is known that (p)ppGpp modulates genes involved in down-regulation of the translational machinery and induction of mechanisms required for stress survival.7, 8 The stringent response also has a role in virulence. 9, 10 It was recently demonstrated in our lab that loss of relA and spoT leads to the increase of PQS and HHQ and increased sensitivity to multiple classes of antibiotics.11
With the emergence of bacteria resistant to most classes of available antibiotics and the shortage of new antimicrobial agents with activity against the Gram-negative organisms, polymyxins (colistin) are considered as one of the last reserve agents for treating Gram-negative bacterial infections. Colistin targets the membrane of Gram-negative bacteria. Electrostatic interactions between the cationic polypeptide (colistin) and the anionic lipopolypeptide (LPS) molecules in the outer membrane of the Gram-negative bacteria leads to a derangement of the cell membrane, which causes an increase in the permeability of the cell envelope, leakage of the cell content, and ultimately causes cell death.12, 13 Cummins et al. demonstrated a striking up-regulation of PQS biosynthesis genes including pqsH, pqsB, and pqsE and the phenazine biosynthesis operon PhzF protein following exposure to sub-inhibitory concentrations of colistin.14 Understanding the exact mechanism on colistin-induced overexpression of PQS biosynthesis genes is critical for the successful development and application of colistin therapy against P. aeruginosa infection.
Jochumsen et al. identified a novel gene, PA5003, that is required for the recognition of CAMPs (Cationic Antimicrobial Peptide) colistin by P. aeruginosa.15 They also showed that PA5003 is also required for the formation of CAMP-tolerant subpopulations in P. aeruginosa hydrodynamic flow chamber biofilms.15 In a random mutagenesis screen conducted in our lab, we demonstrated that loss of PA5003 led to a reduced PQS expression in P. aeruginosa.11 These findings led us to hypothesize that PA5003 is required for colistin-induced PQS overproduction. We tested our hypothesis using P. aeruginosa mutants where PA5003 is inactivated .
The objectives of the study are as follows:
1. To confirm whether sub-inhibitory concentrations of colistin can induce an increased expression of pqsA, one of the PQS biosynthesis genes in P. aeruginosa;
2. To determine whether colistin-induced overexpression of pqsA is through PA5003 in P. aeruginosa.
Materials and Methods
Bacterial strains
The strains used in this study are listed in Table 1.
Table 1. Bacterial strains
Strain name
Genotype & Relevant characteristic
Source or reference
WT
Wild type P. aeruginosa PAO1
Iglewski, B
∆SR
∆relA (∆181-2019) ∆spoT (∆200-1948) unmarked deletion mutant in PAO1 background
(17)
DN287
PAO1 mini-Tn7T-Gm-lacZ
promoterless lacZ reporter
This study
DN288
PAO1 mini-Tn7T-Gm-pqsA-lacZ
wildtype strainwith lacZ transcriptional reporter with pqsA gene promoter
This study
DN296
∆relAspoT mini-Tn7T-Gm-lacZ
∆relAspoT strain with promoterless lacZ without pqsA promoter gene
This study
DN297
∆relAspoT mini-Tn7T-Gm-pqsA-lacZ
∆relAspoT strain with lacZ with pqsA gene promoter.
This study
ΔPA5003
PA5003 transposon mutant with PA5003-G12::ISphoA/hah allele from PW1272, TcR
(16)
Bacterial growth condition: All of the bacteria strains were grown in Mueller Hinton broth at 37 °C and shaking at 250 r.p.m. The overnight cultures were then diluted to an initial OD600 of 0.05 and grown in 15 mL of Mueller Hinton media in 150 mL flasks allowing for optimal aeration and bacterial growth. Colistin was added to a final concentration of either 0.30 µg/mL for PAO1 or 0.15 µg/mL for ΔrelAspoT at time points indicated in experiments.
MIC determination: The test strains were grown overnight on LB agar plates. Several individual colonies were then picked and resuspended in sterile H2O, then vortexed gently until fully resuspended. The dilution 96-wells plate set up by using the micro-broth dilution method, in which the colistin concentration ranges setup from 128 µg/mL to 0.0625 µg/mL. At the same time the corresponding positive controls, drug controls, and media controls were maintained. The plate is then incubated at 37° C for 18 hours. The minimum inhibitory concentration is the lowest concentration of colistin that completely inhibited visible bacterial growth.
β-galactosidaseactivity: The P.aeruginosa transcriptional reporter strains were initially grown overnight in Mueller-Hinton media at 37°C and shaking at 250 r.p.m., Background β-galactosidase activity (promoterless reporter control) was subtracted from all promoter reporter results. β-galactosidase activity was normalized to CFU/ml.
Plasmid DNA isolation: Plasmid DNA was isolated from P. aeruginosa strains using the Compact Prep Plasmid midi kit (Qiagen, Venlo, Netherlands). Bacteria containing plasmids were grown overnight at 37 °C, shaking at 250 r.p.m with the appropriate antibiotic selection (15 mL of Lb, 12.5 µL of ampicillin) for 18 hours. Overnight cultures were centrifuged for 10 minutes at 8000 r.p.m. The supernatant was then discarded and the protocol for plasmid isolation was followed as written in the manual.
DNA transformation: The ΔPA5003 mutant strain was grown in 15 mL of LB in a 150 mL flask overnight. 10 mL of overnight cultures was divided into 1 mL aliquots among 10 sterile 1.5 mL volume microtubes. Aliquoted cells were then centrifuged at 13000 r.p.m. for 2 min. The supernatant was removed and the cells were resuspended in 1 mL of 300 mM sucrose in each tube. This sucrose wash step was repeated once and then all ten cell pellets were resuspended together into a final volume of 400 µl. 1 µg of plasmid DNA was then added to 150 µl of cells. The mixture was then placed in an electrocuvette and electroporated at the following setting (resistance: 200 ohms, capacitance: 125 µFD, 25 µFD, volts: 2.5V). After electroporation, 1 mL of TSB was immediately added to the transformed cell and the cells were incubated at 37°C, shaking at 250 r.p.m. for 2 hours. Cells were then spread out onto selection plates with a concentration of 30 µg/mL and 50 µg/mL of gentamycin. The plates were then incubated overnight and antibiotic resistant colonies were selected and confirmed through PCR of ΔPA5003.
PCR: The polymerase chain reaction was carried out in order to confirm the successful integration of the plasmid into the miniTn7 site of the chromosome. The primers used in this experiment were pGLMS-down (GCACATCGGCGACGTGCTCTC) and PTN7R (CACAGCATAACTGGACTGATTTC). The master piece (Table 2) was first prepared, see the table below. And then adding 24 µL of the master piece and mixed with 1 µL of the DNA that is isolated from the DNeasy Kit protocol and then with the use of the PCR thermocycler programme to amplify the desired DNA, the amplified DNA is then run on the gel (1% of agarose) to check the confirmation of the DNA transformation experiment.
Table 2. Master piece
10X Taq (+KCl, -MgCl2) MgCl2 dNTP (10mM) P1 (61 PTNTR) P2 (16 PGLMS-down) DMSO Taq Ultrapure H2O Data analysis: Prism6 (Graph Pad) was used to produce graphs for each set of experiments as welll as the calculation of means and standard deviation. In addition, we used the student T-test to compare means and determine statistical significance in each experiment.
Results
In order to meet the aims of the project, the first step was to establish the bacterial growth conditions. This was carried out by MIC determination, in which the sub-inhibitory concentration of colistin was established for both PAO1 (0.3 µg/mL) and ∆relAspoT strains (0.15 µg/mL). Figure 1 presents the growth curve of PAO1 and the ∆relAspoT strains under the colistin concentration of 0.3 µg/mL in which both of the strains compared with their controlled condition (no colistin). It appears that there was no significant difference in the growth rate between the PAO1 controlled and the experimental conditions. However, there is a significant difference between the ∆relAspoT strains of the two conditions. This suggested that the concentration of colistin was too high and had an inhibitory effect on the growth of the ∆relAspoT mutant.
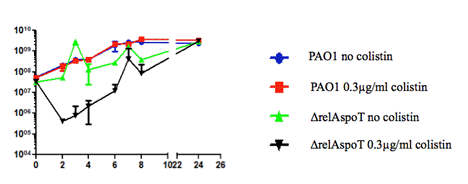
Figure 1. Growth curve of PAO1 and ΔrelAspoT strains under the Muller Hinton Broth with and without colistin (0.3 µg/mL) over 24 hours.
We therefore reduced the concentration of colistin and examined the growth rate again of the ΔrelAspoT strain. Figure 2 presents the growth curve for the ∆relAspoT strain with concentration 0.15 µg/mL of colistin and its control condition. It showed that there was no significant difference of the growth rates between the two conditions, suggesting that the colistin concentration of 0.15 µg/mL was the optimal condition for the ∆relAspoT strain.
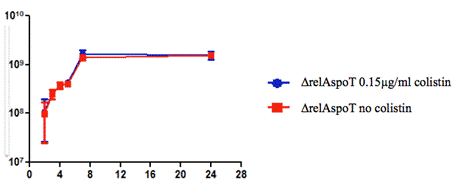
Figure 2. Growth rate of ∆relAspoT strain under concentrations of 0 and 0.15 µg/mL of colistin over 24 hours.
After the optimal growth condition of the bacterial has been established, pqsA-lacZ expression was measured under the optimal conditions using the β-galactosidase assay.
Figure 3 shows the pqsA-lacZ expression of PAO1 strains under the two conditions: one with no colistin and one with 0.3 µg/mL colistin. Colistin induced a statistically significant overexpression of pqsA-LacZ in PAO1 strain at 2 hours, 4 hours, and 6 hours, but this significant over-expression diminished at time point 8 hours and afterwards.
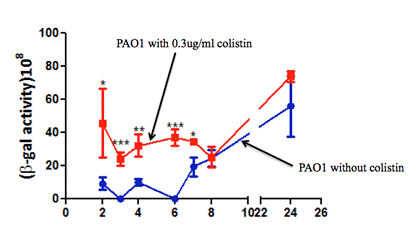
Figure 3. β-galactosidase experiment results of PAO1 strains under two conditions: one with no colistin and one with 0.3 µg/mL colistin. Error bars represent SD. *p<0.05, **p<0.01, ***p<0.001.
Figure 4 presents the pqsA expression of ∆relAspoT strain. Similarly, there was a significant difference in pqsA expression between with and without colistin (0.15 µg/mL) at time point up to 8 hours, and the difference gradually diminished after 8 hours. However, this different over- expression of pqsA was much less in ∆relAspoT strain than that in PAO1 strain. It was also noted that β- galactosidase activity was much higher in ∆relAspoT strain than that in PAO1 strain at 2 hours' time point.
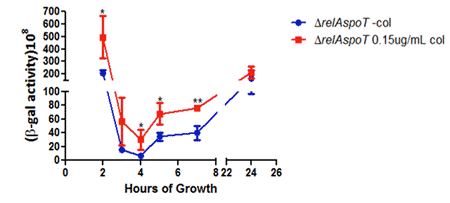
Figure 4. β- galactosidase experiment results of ∆relAspoT strains under two conditions: one with no colistin and one with 0.15 µg/mL colistin. Error bars represent SD. *p<0.05, **p<0.01.
Next, we examined whether colistin can induce over-expression of pqsA in the ∆PA5003 strain.Figure 5 shows that expression of pqsA in both ∆PA5003 strains with and without addition of colistin was reduced over time. Addition of colistin at one hour led to a slightly increased expression of pqsA at time point 3 and 4 hours, but the difference diminished at time point 5 hours and afterwards.
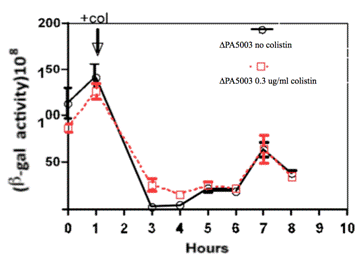
Figure 5. Effects of colistin on pqsA expressions in ∆PA5003. Colistin was added in at time one hour.
Discussion
Colistin is a cationic antimicrobial peptide that targets the bacterial outer membrane by binding to the lipopolysaccharides and displacing the divalent cationic Ca2+ and Mg2+ of the phosphate group leading to the leakage of the intracellular contents and ultimately cell death. It is considered as an important CAMP in the fight against P. aeruginosa infection in CF patients. In order to develop an effective response to the emergence of colistin resistance, it is imperative to understand colistin-induced biological changes in P. aeruginosa.
In the current study, we demonstrated that colistin at a concentration of 0.3 µg/mL can induce a significant increased expression of pqsA in P. aeruginosa PAO1 strains. We also documented that colistin at a concentration of 0.15 µg/mL can induce a significant over-expression of pqsA in P. aeruginosa ∆relAspoT strains. pqsA is one of PQS biosynthesis genes, which regulate numerous virulent compounds production and biofilm formation in P. aeruginosa. Our results suggest that 0.15-0.3 µg/mL concentration of colistin stimulates PQS biosynthesis gene expression and may lead P. aeruginosa to adaptive responses to colistin therapy. A previous study by Cummins et al.14 demonstrated the sub-inhibitory concentrations of the colistin induce striking up-regulation of production of PQS biosynthesis genes, including pqsH, pqsB, and pqsE and the phenazine biosynthesis operon. Our findings are consistent with Cummins's and added further evidence that pqsA can also be upregulated by sub-inhibitory concentration of colistin.
Further, we documented that expression of pqsA is reduced in PA5003 mutant strains. Treatment of colistin in ∆PA5003 strains only induces a minimal increase in pqsA expression at 3 and 4 hours but diminished afterwards. Our findings suggest that colistin is likely to induce over-expression of PQS biosynthesis genes via PA5003. Further studies are needed to confirm these results.
Conclusion
We confirmed that sub-inhibitory concentrations of colistin can induce over-expression of PQS biosynthesis genes in P. aeruginosa and demonstrated that this inducing might be via PA5003. Although further studies are needed to confirm the results, our findings provide new insight into developing an effective colistin therapy in the fight against P. aeruginosa infections in CF patients.
Acknowledgement
The project is supported by the Burroughs Wellcome Fund to Dr. Dao Nguyen.
Footnotes
1. Yankaskas, J.R., Marshall, B.C., Sufian, B., Simon, R.H. & Rodman, D. Cystic fibrosis adult care: consensus conference report. Chest 125, 1S-39S (2004).
2. Spilker, T., Coenye, T., Vandamme, P. & LiPuma, J.J. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol 42, 2074-9 (2004).
3. Deziel, E. et al. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101, 1339-44 (2004).
4. Haussler, S. & Becker, T. The pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog 4, e1000166 (2008).
5. Pesci, E.C. et al. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96, 11229-34 (1999).
6. Potrykus, K. & Cashel, M. (p)ppGpp: still magical? Annu Rev Microbiol 62, 35-51 (2008).
7. Vogt, S.L. et al. The stringent response is essential for Pseudomonas aeruginosa virulence in the rat lung agar bead and Drosophila melanogaster feeding models of infection. Infect Immun 79, 4094-104 (2011).
8. Erickson, D.L., Lines, J.L., Pesci, E.C., Venturi, V. & Storey, D.G. Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster. Infect Immun 72, 5638-45 (2004).
9. Falagas, M.E. & Kasiakou, S.K. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis 40, 1333-41 (2005).
10. Davis, S.D., Iannetta, A. & Wedgwood, R.J. Activity of colistin against Pseudomonas aeruginosa: inhibition by calcium. J Infect Dis 124, 610-2 (1971).
11. Schafhauser, J. et al. The stringent response modulates 4-hydroxy-2-alkylquinoline biosynthesis and quorum-sensing hierarchy in Pseudomonas aeruginosa. J Bacteriol 196, 1641-50 (2014).
12. Schindler, M. & Osborn, M.J. Interaction of divalent cations and polymyxin B with lipopolysaccharide. Biochemistry 18, 4425-30 (1979).
13. Sabuda, D.M. et al. Utilization of colistin for treatment of multidrug-resistant Pseudomonas aeruginosa. Can J Infect Dis Med Microbiol 19, 413-8 (2008).
14. Cummins, J., Reen, F.J., Baysse, C., Mooij, M.J. & O'Gara, F. Subinhibitory concentrations of the cationic antimicrobial peptide colistin induce the pseudomonas quinolone signal in Pseudomonas aeruginosa. Microbiology 155, 2826-37 (2009).
15. Jochumsen, N., Liu, Y., Molin, S. & Folkesson, A. A Mig-14-like protein (PA5003) affects antimicrobial peptide recognition in Pseudomonas aeruginosa. Microbiology 157, 2647-57 (2011).
16. Held K, Ramage E, Jacobs M, Gallagher L, Manoil C. Sequence-Verified Two-Allele Transposon Mutant Library for Pseudomonas aeruginosa PAO1. Journal of Bacteriology. 2012;194(23):6387-9
17. Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, et al. Active Starvation Responses Mediate Antibiotic Tolerance in BIofilms and Nutrient-Limited Bacteria. Science. 2011;334(6058):982-6.
|